It’s that time of year in Boston when the flowers bloom, the trees bud and the protein science community emerges from labs across the country to make the migration, long or short, to PEGS Boston.
This annual meeting of the minds, regarded by many as the “go-to event” for biologics and protein engineering, convenes global leaders and researchers to share new research and fresh ideas. According to André Teixeira, senior director of the antibody platform at the Institute for Protein Innovation (IPI), “the best thing about PEGS is you see what people are working on and where the future is.”
This year, IPI had plenty to add to the conversation.
Scientist Rebecca Hershman regaled listeners with a colorful account of IPI’s antibody discovery journey, a holistic soup-to-nuts process that passes from antigen generation to characterization. Her poster focused on how the IPI team successfully sifted through a pool of millions of contenders to find a promising cluster of anti-semaphorins, which are now entering larger-scale production at IPI. Hershman was also awed by the different modalities being generated for multi-specifics and added valencies — “there’s a lot of creativity there,” she said.
A Danish chair is to a Baroque throne what IPI’s antibody library is to traditional display techniques — sleek, minimalist and designed for functionality. In his poster, senior scientist Deepash Kothiwal shared the craftsmanship involved in the IPI yeast display library and our elegant approach to developing high-affinity Fabs and VHH antibodies. He left the conference mulling new trends in discovery and engineering. With the rapid integration of artificial intelligence (AI) and emerging therapeutic areas, scientists will need to “remain adaptable and continuously update their skill sets,” Kothiwal said.
Curtis Walton, director of automation, showed off the bespoke, flexible automated workstation used for antibody discovery at IPI. In the past year, Walton has revolutionized IPI’s workflow by integrating hardware and software updates throughout the pipeline, focusing first on the Discovery Team to speed antibody selection output. But the ever-curious Walton found inspiration in work presented by Merck, Bayer, Johnson & Johnson, Adaptyv Biosystems and AstraZeneca. “It was great to see the innovative ways teams have built platforms to accelerate antibody discovery and biotherapeutic development,” he said.

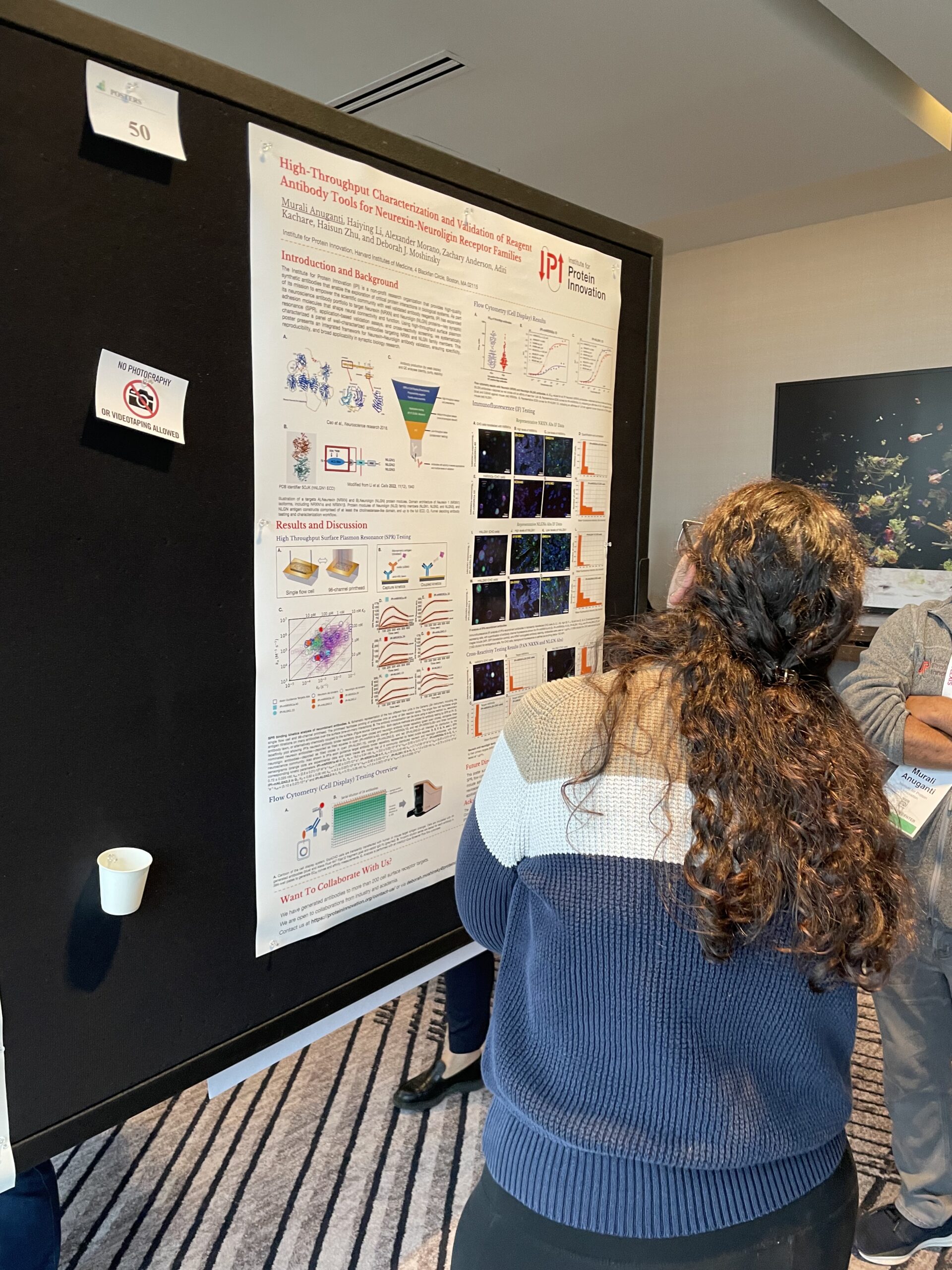
IPI is determined to develop reliable recombinants that change the game of biomedical research — but if ensuring that reliability was easy, everyone would do it. Using IPI’s neurexin and neuroligin antibodies as a case study, senior scientist Murali Anuganti detailed our in-house approach to characterization and validation. The team puts surface plasmon resonance, validation assays and cross-reactivity screening to work to evidence specificity, reproducibility and broad applicability in synaptic biology — three hallmarks of quality and reliability. “It’s not just about having a great antibody—it’s about proving it works across the right species, tissues, and applications,” Anuganti said.
Rob Meijers, head of IPI’s Neuroscience Program, took the opportunity to take the PEGS audience through some of the lessons learned over the last years from high throughput production of glycoprotein antigens. IPI has developed a trimmed down and efficient plasmid for transient mammalian glycoprotein production. The pTipi vector is now available from Addgene and the Belgian Coordinated Collections of Microorganisms (BCCM) through a no-frills license. Meijers also presented a study on codon optimization schemes, showing some surprising trends that now inform antigen production at IPI and resonated well with the PEGS audience, leading to a spirited discussion. This was done using a high-throughput ELISA fluorescence approach capable of rapidly assessing multiple recombinant constructs simultaneously.

Senior scientist Anita Ghosh has a favorite tag line for deploying protein targets in antibody discovery — “garbage in, garbage out.” Good output is not guaranteed unless discovery platforms take the time to produce and purify recombinant native mammalian proteins in microgram to milligram quantities. But this early diligence often creates bottlenecks, so Ghosh and IPI have optimized the process to achieve high-throughput-like efficiency that keeps pace with the rest of the pipeline. She presented this nifty approach at PEGS 2025 and the crowd went wild (as much as a room of bioscientists in uncomfortable chairs can).
Seems like everyone is talking about machine learning and AI these days, and IPI is no different. Teixeira showcased how this wizardry could help IPI develop a small, pipeline-specific model that streamlines discovery by predicting biophysical properties and binding capabilities. In a collaborative computational science, antibody discovery and antibody engineering effort, the team fed high-throughput data from the IPI platform — including data on yeast display, sequencing and characterization — into a protein language model to reveal the most promising binders. “We are not using AI just for the sake of it, but we have identified problems that could potentially be better solved by using the technology,” Teixeira said. “I think a lot of people can relate to that, and we got some pretty good feedback during the conference.”
Sources:
Rebecca Hershman
Deepash Kothiwal
Curtis Walton
Murali Anuganti
Anita Ghosh
André Teixeira
Rob Meijers
Writer:
Caitlin Faulds, caitlin.faulds@proteininnovation.org
About IPI
The Institute for Protein Innovation is pioneering a new approach to scientific discovery and collaboration. As a nonprofit research institute, we provide the biomedical research community with synthetic antibodies and deep protein expertise, empowering scientists to explore fundamental biological processes and pinpoint new targets for therapeutic development. Our mission is to advance protein science to accelerate research and improve human health. For more information, visit proteininnovation.org or follow us on social media, @ipiproteins.


