IPI epitope tag antibodies, delivered via an open science model, aim to increase access, cut costs and enhance reproducibility.
Maybe this story is yours. You’ve got a protein you want to detect in a particular application and species. You can’t find a good antibody; those available aren’t specific, cross-react or just don’t work. Or maybe you are working with a novel protein that doesn’t yet have a good reagent binder. You’ve searched all the catalogs and asked everyone you knew in the field. In the end, you just can’t come up with a solution that works.
What do you do?
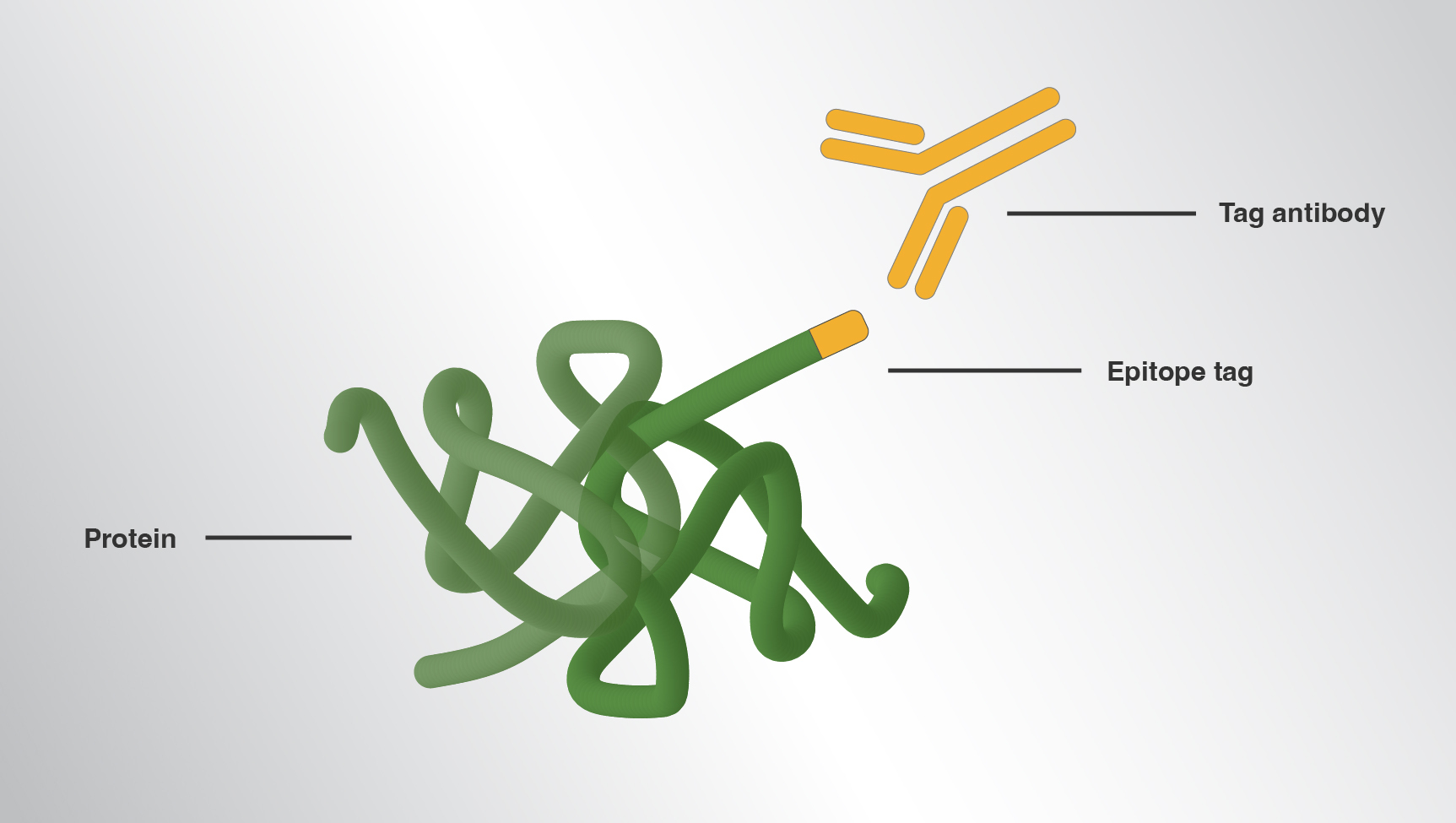
You can try epitope tags — short amino acid sequences fused to your protein’s N- or C-terminus. When paired with an antibody that specifically and reproducibly recognizes the tag, you can purify, detect or localize your protein, zero in on its function and probe its hundreds of thousands of potential interactions.
To fill a gap in easily accessible protein tools, the Institute for Protein Innovation (IPI) and its partner, Addgene, have developed inexpensive antibodies targeting common epitope tags. The antibodies are now available, as part of IPI’s new catalog of recombinant antibodies and reagents. Soon after, IPI will release the plasmids that encode the antibodies.
In addition to the antibodies and plasmids themselves, “we are making the sequences available using an open science model,” says Rob Meijers, senior director of the antibody platform at IPI. “That allows people to make the antibody in their own lab, save a lot of money and have control over the material.”
A valuable protein tool
Epitope tags have long proven valuable for researchers working with novel, poorly immunogenic or scantly expressed proteins. Tags are also commonly used in a variety of applications including immunoprecipitation and super-resolution microscopy. When antibodies against a target protein are lacking, scientists generally tag their protein and immunoprecipitate the construct with the corresponding tag antibody.
“Anyone who’s working with proteins is probably using a tag,” says Meghan Rego, director of antibodies at Addgene.
Researchers typically purchase V5, c-Myc, HA or one of a dozen other types of epitope tag antibodies by the microgram or milligram from reagent antibody companies. But the cost of currently available tags is high and can add up to thousands of dollars for just a single tag antibody.
Often, scientists developing therapeutics can justify the cost. Basic scientists often cannot.
“Researchers at institutions that aren’t as well-funded might not be able to afford to order these antibodies from these companies,” Rego says. “Or they might have a hard time shipping something perishable across the world. So, for them, having a sequence makes it a lot easier.”
Another issue is reproducibility. In a 2015 Nature commentary, more than 100 researchers pointed to “serious flaws” in the reliability of “poorly characterized and ill-defined antibodies.”
In 2023, Carl Laflamme at the Structural Genomics Consortium and colleagues reported that in a comprehensive third-party test of 614 commercial antibodies, only around a third of polyclonal and monoclonal antibodies recognized their target in the experimental approaches they were recommended for.
“IPI is taking away that ambiguity,” Rego says. “If someone needed to repeat your paper 40 years from now, they don’t have to worry about whether or not that company still exists or if that antibody has been lost.”
By making the antibody sequence available, IPI and Addgene are providing researchers with a means to produce an exact version at any time.
Epitope tags in use
The molecular toolbox of tags is an alphabet soup of names: V5, Protein C, DYKDDDDK, Rho, His, Biotin, EE, GCN4, MBP and Strep, among others. The choice of tag comes down to which is best suited for your particular assay.
For example, the 14-amino-acid V5 tag, derived from simian virus 5 (SV5), provides a reliable way to detect target proteins. Meijers notes that IPI commonly uses the V5 tag in its own antibody discovery operation.
Specifically, IPI researchers fuse the V5 tag to antibody sequences and transfect the constructs into yeast cells. The microorganisms express the resulting tag and antibody on their surfaces.
“Then we can see which cells are displaying the antibody (via its tag),” Meijers says, “and get rid of the cells that don’t display it.”
Similarly, tagging a protein with tags like V5 allows Rego to see it on a Western blot. She can easily remove the tag afterward to ensure it will not affect the protein’s future behavior.
A note of caution: some tags can interfere with protein structure, activity or binding affinity. It’s essential to investigate before going down the epitope tag route.
A long-lasting collection
To create its collection of tag antibodies, IPI took advantage of the fact that many of the original epitope tags are now freely available off-patent. The team fused the variable sequences of the original antibodies to the constant regions of a rabbit IgG. This rabbit framework allows researchers to use the same secondary antibody (an antibody that recognizes the first antibody) to detect all the epitope tags in the IPI catalog.
“We converted the originals into recombinant epitope tag antibodies,” Meijers says. “Now, they are forever renewable.”
Source: Rob Meijers, rob.meijers@proteininnovation.org
Meghan Rego, meghan@addgene.org
Writer: Deborah Halber, dhalber@writingreality.com
About IPI
The Institute for Protein Innovation is pioneering a new approach to scientific discovery and collaboration. As a nonprofit research institute, we provide the biomedical research community with synthetic antibodies and deep protein expertise, empowering scientists to explore fundamental biological processes and pinpoint new targets for therapeutic development. Our mission is to advance protein science to accelerate research and improve human health. For more information, visit proteininnovation.org or follow us on social media, @ipiproteins.